Discussion on coronavirus variants have been the rage of the town in the COVID-19 pandemic. Surabhi Srivastava, Research Coordinator, Genomics and Bioinformatics at CCMB has been at the forefront of studying these variants. She takes us through the evolution of the virus, explaining the details of the process, the current scene in India and the way forward.
In navigating the second year of the COVID-19 pandemic caused by the SARS-CoV-2 coronavirus, India was caught by a brutal summer wave of infections. Beginning March 2021, the country saw a steep rise in case numbers, making for a grim total of 3.3 crore cases and well over 4 lakh deaths so far. This was accompanied by an increase in the emergence and spread of new forms of SARS-CoV-2 such as the Delta and the so-called Delta plus variants.
Along with the genomics team at the Centre for Cellular and Molecular Biology (CSIR-CCMB), I have been monitoring the spread of the virus since the beginning of the pandemic and closely following its evolution. We have seen how we can be repeatedly blindsided as the virus rages in different forms from one part of the globe to the next. In this article, I highlight how SARS-CoV-2 changes to give rise to new variants, their biological significance, and how we can do our part in stopping the emergence of newer variants.
Essence of SARS-CoV-2 genome
Ribonucleic Acid or RNA, a single strand of ~30,000 bases, makes up all the genes of SARS-CoV-2 – the viral genome. These genes code for the viral proteins that make its structure and perform the functions needed for it to infect host cells and survive. As the virus multiplies inside human cells, it also makes copies of its genome for the new viral particles. Over time, copying errors are introduced in the viral genome due to the error-prone RNA-copying machinery. These errors are called mutations and can lead to changes in the encoded viral proteins.
Most of these changes are of no consequence and don’t even get noticed. But some of them can change the infectivity, transmission and/or other features and get selected for their beneficial properties to the virus. Over the past year and a half of the pandemic, SARS-CoV-2 has been accumulating mutations at the rate of ~2 changes per month.
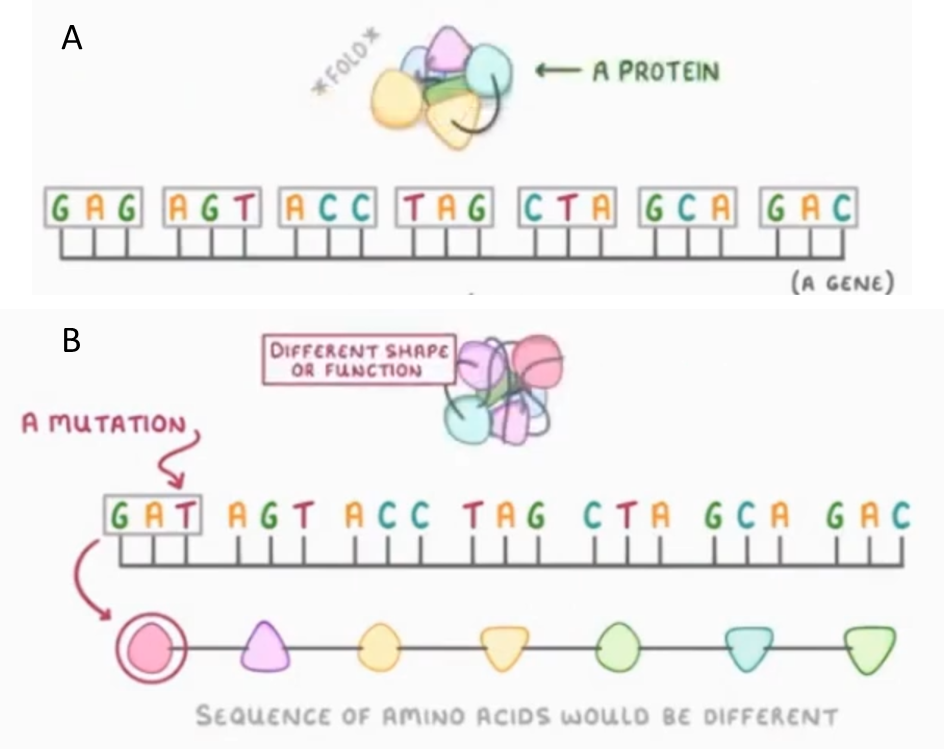
Figure 1: Mutations alter genes and their encoded proteins. A) The genomic sequence of the bases – Guanine (G), Cytosine (C), Adenine (A) and Thymine (T) in DNA/Uracil (U) in RNA – defines a gene, which encodes a protein made up of amino acids. B) Mutations or a change in the genomic sequence causes a change in the amino acid sequence and the encoded protein. Figure adapted from BIC, CSIR-CCMB.
Variants of Interest and Concern
Variants of Interest (VOIs) carry one or mutations that are believed to have biological consequences with harmful effects for the human hosts. It is important to note that more mutations do not automatically mean a more dangerous variant; it depends on the type and combination of mutations. Typically, any VOI has a constellation of multiple mutations, and sometimes one or more of these help in improving viral binding to the human receptor, easing its entry into human cells, evading the body’s immune response or escaping current diagnostic strategies used to identify the infection.
Early this year the WHO created a fresh blueprint to look into such mutations and identify emerging and potentially dangerous variants. Experimental and epidemiological studies identify the role of the specific mutations in a particular VOI to find evidence for increased transmission, virulence or immune escape. It then becomes a ‘Variant of Concern’ (VOC) and its spread is monitored. As the variant transmission increases, its proportion in the virus population rises to form a recognizable lineage – a set of viral variants sharing a common ancestor. Once the representative number crosses a threshold, the lineage is assigned a name and the variant should be referred to by its official name.
In common usage, however, new variants have been named by the country where they were first found. The WHO recently devised a new naming scheme for VOIs and VOCs based on the letters of the Greek alphabet, which are easier to remember and non-stigmatizing to countries.
Genome surveillance of SARS-CoV-2 in India
New viral variants keep emerging. Next generation sequencing (NGS) technologies allow researchers to decipher the order and arrangement of the bases in a genome of a viral lineage, a process termed as genome surveillance.
Viral genome surveillance enables monitoring of the SARS-CoV-2 genome sequence to identify mutations arising in the strains infecting population of a certain region. It is crucial for tracking the effects of emerging mutations. However, carrying out genomic surveillance in a populous country like India requires rapid and coordinated temporal sequencing of viral genomes across multiple states.
The Indian SARS-CoV-2 Genome Sequencing Consortia (INSACOG) was formed towards the end of 2020. Its primary aim is to identify the emergence and spread of more transmissible VOCs such as those identified nationally as well as in the UK, South Africa and Brazil. In the longer scheme of things, the consortium plans to sequence a fraction of all positive cases identified in the country through the duration of the pandemic. Over 50,000 viral genome sequences from India are currently available in the global public database called Global Initiative on Sharing All Influenza Data (GISAID) to enable data-sharing among researchers.
India has state-of-the-art high throughput genome sequencing and data analysis facilities at multiple labs contributing to the INSACOG initiative. 28 labs from the Ministry of Health, Department of Biotechnology (DBT), Indian Council of Medical Research (ICMR) and Council of Scientific and Industrial Research (CSIR) are involved in large-scale genome surveillance endeavors across the country. This has been possible due to the large network developed with major hospitals and COVID-19 testing centres as well as state governments over the last year, for access to patient samples and epidemiological data. However, given the country’s population and COVID-19 case burden, it is imperative to incorporate further measures to upscale genomic surveillance programmes across the country.
CSIR-CCMB has been spearheading the COVID-19 testing and sequencing of viral samples from infected cases, especially across Telangana and Andhra Pradesh. We have sequenced nearly 8000 viral genomes and analyzed viral variants circulating across the country to follow emergent mutation trends. The sequences and mutations analysis are hosted on our interactive dashboard, GEAR-19 (Genome Evolution Analysis Resource for COVID-19), with an easy graphical user interface so that the data is publicly available to researches across the country and the globe.
Identifying concerns in current VOCs
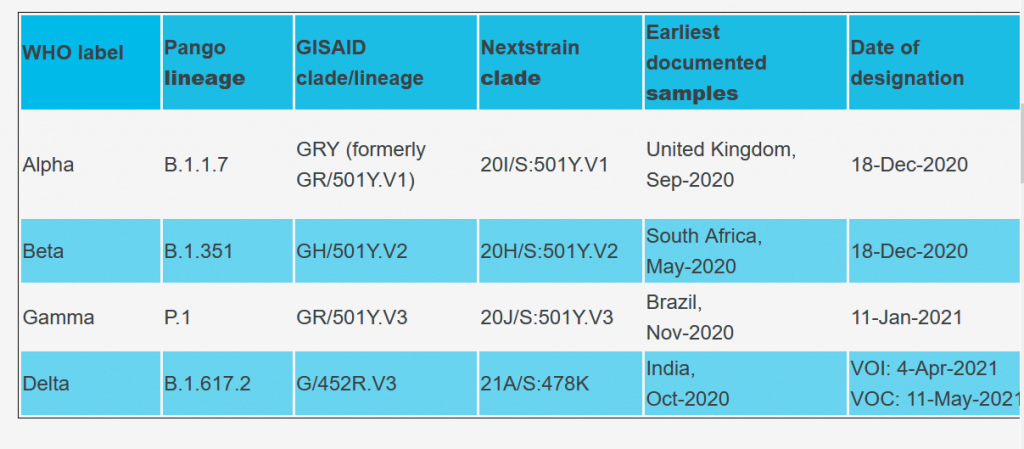
There are currently four major VOCs listed by the WHO (Table 1).
Table 1: List of current VoCs with their multiple nomenclature
The Delta variant has been the latest addition to the WHO watchlist, though its origins in India are traced much earlier. This arose in Maharashtra as a cluster dubbed ‘double mutant’ early in 2021. The surveillance efforts marked it concerning because of its two mutations – L452R and E484Q – in the viral Spike gene. Both mutations were known from other VOIs that had shown evidence for concern earlier.
The L452R mutation alters the Spike protein on the viral surface that facilitates binding of the virus to human cells. This change can facilitate enhanced binding of the virus to human cells, and possibly weaken neutralization of the virus. The E484Q mutation changes the antibody recognition site on the virus. As a result, antibodies developed by a prior coronavirus infection or vaccination might not work as well, allowing viral immune escape.
Multiple independent mutations at the position 484 have been seen earlier in other variants too, including the Beta and Gamma variants. This suggests mutation at this position possibly offers a selective advantage to the virus. N501Y is another mutation that offers advantage to the virus by enhancing its transmissibility. In addition to the Beta and Gamma, this mutation is seen in Alpha as well.
The lineage from India, on the other hand, bears the P681R mutation and was flagged for concern by the Indian government in March 2021. Subsequently, the double mutant was officially named B.1.617 lineage. It has evolved into three sub-lineages since then. One of these – variant B.1.617.2 – acquired very high transmission capability compared to the ancestral strain and has risen to global prominence, spreading to nearly 120 countries (https://outbreak.info/). It was declared a VOC by the World Health Organization (WHO) and designated as the Delta variant in May 2021.
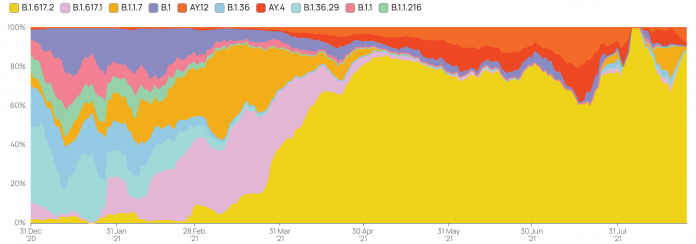
Recent studies have indicated viral loads over 1,000 times higher in people infected with the Delta variant than those infected with the earlier coronavirus strains. The incubation period is also shortened upon infection with the Delta variant, contributing to its high rate of transmission and spread. During the second wave of COVID-19 in India, the Delta variant became the predominant SARS-CoV-2 lineage and is associated with the steep increase in cases seen in the country during the summer. The rapid rise of Delta variant in India is highlighted in figure 2 (B.1.617.2, green color), although other variants can still be found in various pockets of the country.
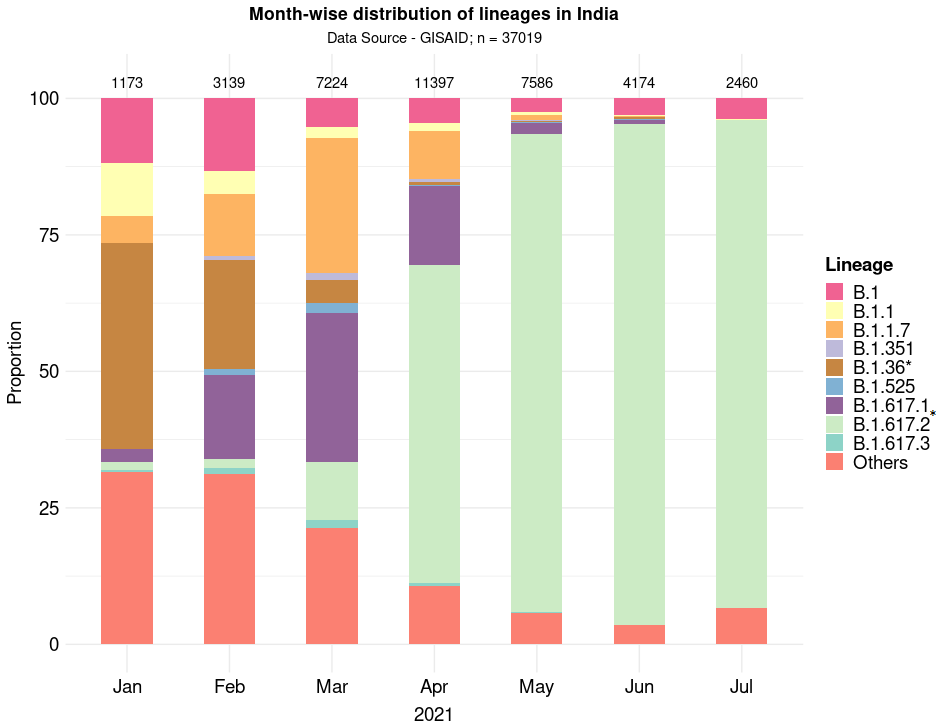
Figure 2: SARS-CoV-2 variants in India in 2021. The Y-axis shows the proportion of top variants (color coded) over the last 7 months (X-axis). *includes sub-lineages. Plot: Sofia Banu, BIC-CCMB. Data source: GISAID
The road ahead
The only means of replication available to SARS-CoV-2 is inside its human host. Higher disease spread and increasing infections, therefore, provide greater avenues for the SARS-CoV-2 genome to develop multiple mutations, leading to new variants of the virus. Not all mutations manage to obtain a stronghold in a population. The more transmissible variants can quickly wipe out other lineages despite the presence of concerning mutations. A case in point in the Indian context is the N440K mutation (in B.1.36 and its sub-lineages). Associated with potential immune escape, it was seen to be rising prior to the second wave in the country (Figure 2). But it was overtaken by the Alpha and then by the Delta variant.
Over the last few months, Delta has continued to evolve into the colloquially dubbed Delta plus and beyond, and now has many sub-lineages designated as the AY series. The family comprises over 30 global variants; each of these carries additional mutations/combinations on the Delta background. These new variants are epidemiologically relevant but do not seem to have acquired any clinical advantage over Delta so far.
Researchers are also keeping an eye out for other mutations that might confer specific advantages to the SARS-CoV-2. These can evolve multiple times independently at various locations – a phenomenon called convergent evolution – and are tracked by genomic analysis. Though they can dramatically change the viral transmission dynamics, variants alone do not govern the course of a pandemic. Besides the mutation profile and the advantage conferred to the virus, the control measures being undertaken at the time and place of emergence, degree of travel and vaccination status together determine the eventual dominance and spread of a new variant. Hence, real time epidemiological analysis needs to complement genomic surveillance in order to catch a dangerous variant as it emerges and guide public health measures to counteract it.
Finally, there is a collective responsibility that we all hold in order to stay on top of this pandemic. Avoiding large and indoor gatherings and promoting the use of well-ventilated spaces has a huge impact in slowing community transmission by depriving the virus of new host populations. Measures like physical distancing, correct masking and sanitization dampen the evolution and spread of all variants and will be crucial till the pandemic subsides and/or SARS-CoV-2 becomes endemic. The unprecedented success in the development of COVID-19 vaccines has given the world a chance to control the pandemic with minimum loss of life. Without adequate change in our behavior and widespread adoption of these interventions, conditions favor the emergence of new variants that will challenge the edge provided by the vaccines.