Cover art: Genome connections by Dr Sweena Chaudhari Joshi
A couple of months ago, I came across an article, about how scientists had recreated the faces of Egyptian mummies from just their DNA! They were able to generate the minor details of their faces, the light brown eyes, and the freckles. I was very fascinated and read it multiple times. It explained how scientists had recreated the 3D models of mummies’ faces using ‘genome sequencing’. But I, a pure mathematics student, had no idea what that meant.
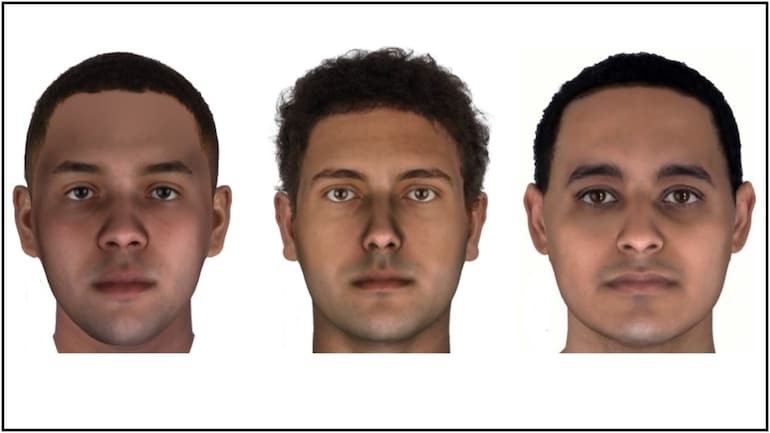
I reached out to some scientists at the Centre for Cellular and Molecular Biology (CCMB) who perform genome sequencing regularly and analyze the results regularly. I spent time with them trying to understand ‘Next Generation Sequencing’ and its different uses!
What is a genome? What is sequencing? What is next-generation sequencing? If these questions bother you too, as they did me, this article is for you.
Let’s start with the genome.
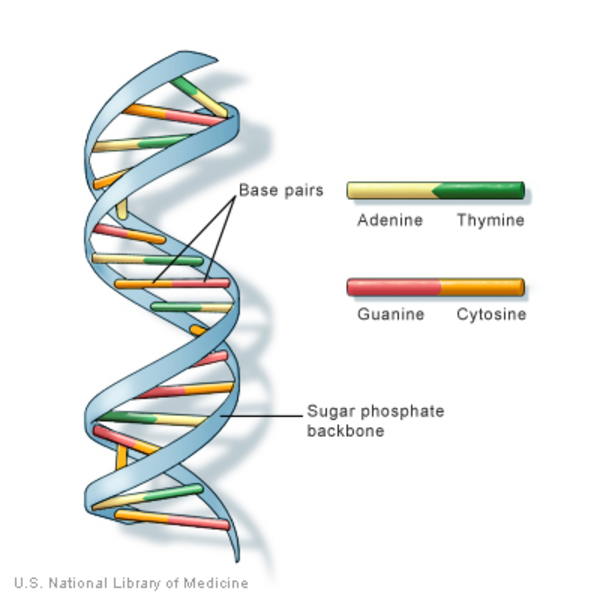
A genome is an organism’s full set of genetic instructions (genetic code). It contains all the information needed to build and develop that organism and for the organism to function. The instructions in a genome are made of a complex molecule called DNA (deoxyribonucleic acid). DNA is a double helix structure composed of smaller chemical molecules called nucleotide bases: adenine (A), cytosine (C), guanine (G), and thymine (T). The base A on one of the strands is paired with T and G with C on the other strand. The backbones of the helices are made up of sugar and phosphate molecules that are connected by chemical bonds.
Sections of this DNA make up genes. The composition and sequence of nucleotide bases in genes control a lot of aspects in our bodies – color of our eyes, height, anger – the list is long and keeps growing. How do genes control all these features? Genes contain information to make a class of molecules called proteins in our cells. This happens through a complex 2-step process called gene expression. Simply put, genes make RNA (ribonucleic acid) and RNA makes proteins. Proteins are large and complex molecules that perform various important functions in our body. It is the proteins that are responsible for your green eyes and tall height. Changes in the order of nucleotide bases in the genes can, thus, affect the proteins and the biological processes they control.
There is a total of 3.2 billion nucleotides in the human genome. Humans and chimpanzee genomes are ~99% similar. Among two individual humans, the similarity is much higher up to ~99.9%. This 0.1% difference in bases between two humans accounts for our unique features as well as our susceptibility to various diseases!
Now that we know what a genome is and what it is composed of, we move on to our next question. What is sequencing?
In simple words, sequencing is the ability to read the genetic code. It is a laboratory technique used to determine the exact sequence of bases in a DNA molecule. Knowing the exact sequence of the bases helps us to understand the functions of the gene and predict the structure and function of a protein that it encodes.
An extremely popular example of sequencing is the “Human Genome Project.” Its objective was to determine the entire DNA sequence of the human genome. In 2003, the first accurate and complete human genome sequence was finished two years ahead of schedule. It took 13 years to complete this project! Technology has improved a lot since then and we can now sequence hundreds and thousands of genes or a whole human genome in a couple of days! Next-Generation Sequencing (NGS) is one such technology that has enabled us to determine the order of nucleotides in entire genomes or targeted regions of DNA or RNA.
What is special about NGS?
NGS is a technology used to determine the sequence of nucleotides making an RNA or DNA. Broadly, scientists extract RNA/DNA from cells and purify it from the other debris in the cell. They slice out the parts of it that are of interest to them and put it in a sequencer machine to read its nucleotide composition. Some machines identify the bases based on their sizes and shapes. Some open the two helices of DNA, use pre-labeled fluorescent nucleotides that only bind to the complementary partner, and can keep a track of the binding nucleotides.
I got to learn about many interesting applications of NGS, which can be used to study various biological phenomena. Some of those are:
- Genome / Exome sequencing:
Genome sequencing is a process where we try to determine the exact sequence of the nucleotide bases of the entire DNA strand. On the other hand, exome sequencing, also known as whole-exome sequencing, is a technique for sequencing only the protein-coding regions of genes in a genome (the exome). It is a 2-step process in which the protein-coding regions are first identified and then selectively sequenced using DNA sequencing technology.
In the case of human diseases, scientists can compare the DNA sample of affected individuals with the reference genome or exome of a non-diseased human and figure out the difference in the sequences. These changes in sequence could be a substitution, deletion, or addition of a base, and are called mutations. Any minor change in the nucleotide base sequence can alter the encoded protein and this can be sometimes enough to cause disease.
Not just for humans, but genomes can be sequenced and compared for other species as well. One of the most relevant examples in current times is the sequencing of SARS-CoV-2 genomes – the virus that causes COVID-19. By sequencing the entire viral genome, scientists can look at changes or mutations in viruses infecting different people. Since proteins govern all vital functions, some of these mutations can change the effect of the viral proteins to give an advantage to the virus in terms of increased transmission or infectivity in human hosts. And that can have consequences for pandemic management in terms of the speed with which the virus spreads or the severity of the disease. Therefore, this has been a critical aspect of the world’s pandemic surveillance program and India is no exception. I learned that CCMB has been involved in viral genome surveillance since the very beginning in March 2020 and their analysis can be found on GEAR-19.
- RNA/Transcriptome sequencing:
All the RNA expressed from genes in a cell is called the transcriptome. It governs the nature and amount of functional protein to be made in a cell. So, scientists also study the functioning of a gene by sequencing the RNA made, and how it differs under various conditions such as stress or diseases.
In the context of the pandemic, scientists at CCMB sequenced the transcriptomes of Indian patients suffering from COVID-19. They found that the RNA profile or the transcriptome of these patients was different from uninfected people because a lot of the immune response genes got activated upon viral infection. This makes sense as the body’s mechanism is triggered to fight the infection but studying which RNAs were being highly produced helps understand and even predict the severity of the infection in the patient.
- Metagenomics:
Metagenomics is the study of multiple genomes representing a diverse community of organisms. Often it involves genetic material recovered directly from environmental samples, such as soil, air, or water, that contain many microorganisms. Metagenomics is, therefore, used to study the microbial diversity and ecology of a specific environment. And wait, this environment can also be our gut! Scientists have studied the genomes of the many microbes that house in our gut and suggest their roles in health and disease.
There are some very interesting studies planned to establish the gut microbiome of Indians. Given our high ethnic and geographical diversity and the variety of dietary habits of different groups of individuals in the country, it is important to establish the baseline of different microbiomes across our people. And Indian scientists, including at CCMB, also want to study how these gut microbiomes interact with our genomes, and it that affect our inherited propensity towards different diseases. Sometime in the future of people may have their personal genome cards containing all this information, with their susceptibilities to diseases and their cures already known!
- Genome Assembly:
There are many organisms whose reference genomes are not yet generated or sometimes, scientists discover new organisms whose genomes are unknown. To study the evolution and characteristics of these species, scientists use a process called “de novo genome assembly” to create a reference genome of an organism. Assembly is a process where short sequences of nucleotides (acquired by cleaving the larger DNA strands at specific sites by restriction enzymes) are put together to create a longer and larger fragment of the genome. Using the various computational tools of de novo genome assembly, scientists can sequence the DNA and ‘build’ the reference genome of any organism.
So far, only 0.1% of animal and plant species of DNA has been sequenced. Increasing our understanding of Earth’s biodiversity and responsibly stewarding its resources are among the most crucial scientific and social challenges of the new millennium. The Earth Bio Genome Project: Sequencing life for the future of life (EBP), launched on 1st November 2018 hopes to tackle this challenge. The project aims to sequence, catalog, and characterize the genomes of nearly 1.5 million known animals, plants, protozoans, and fungal species on earth over a period of the next 10 years at an approximate cost of US$ 4.7 billion.
The outcomes of the EBP will inform a broad range of major issues facing humanity, such as the impact of climate change on biodiversity, the conservation of endangered species and ecosystems, the preservation and enhancement of ecosystem services, and most importantly, will lead to the generation of the genetic blueprint of all living forms. India also aims to participate in this global initiative, with the team at CCMB hoping to participate in sequencing 1000 unique species of relevance to the country in the next 5 years! And these will require an equally adept Bioinformatics and Data Analytics team to analyze the sequencing results and make meaningful and visually easy-to-understand inferences.
So, in the coming years, NGS tools are positioned to solve the ancient mysteries in history as well as inform a broad range of major issues facing humanity. From uncovering the Egyptian mummies to understanding disease prevalence and ushering in the era of personalized medicine, from studying the impact of climate change on biodiversity to the conservation of endangered species and ecosystems – NGS is here to stay.
Acknowledgments: Many thanks to Dr. Surabhi Srivastava for her constant inputs and edits in this article. Dr Srivastava is the Genomics coordinator at CCMB.


